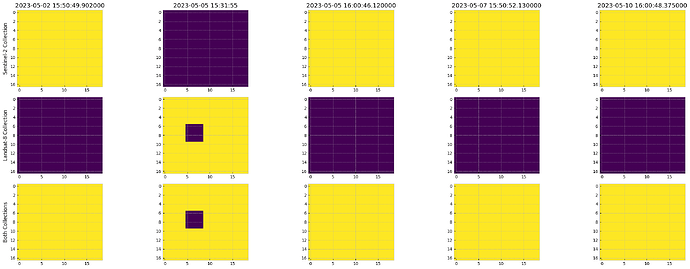
Thank you, @william.ray. I ran it again with your evalscrip. When I run it with the date range 2023-05-07- 2023-5-10 with “constellation”: “LANDSAT” I have two images returned and when using “SENTINEL” no images are returned. The problem is that for the time period I used, there were no Landsat images. I shouldn’t have any images returned when I specify “LANDSAT” but instead I get two Sentinel images. It appears as if when all the “bands” in the evalscript exist for both Landsat and Sentinel then the output results will be the same regardless if “LANDSAT” or “SENTINEL” are requested. If, as in your evalscrip, one or more bands do not exist in the specified collection, then no images are returned for that collection. Below is the script I’m using. Here are the image dates and the associated satellite (S30 or L30) in the date range specified in the script. I verified that these images exist in the Sentinel Hub archive and the NASA archive.
HLS.S30.T18TXQ.2023130T154819.v2.0
HLS.S30.T18TXQ.2023127T153819.v2.0
HLS.S30.T18TXQ.2023125T154811.v2.0
HLS.L30.T18TXQ.2023125T153155.v2.0
HLS.S30.T18TXQ.2023122T153811.v2.0
from sentinelhub import CRS, BBox, DataCollection, SHConfig
from eolearn.core import EOWorkflow, FeatureType, LoadTask, OutputTask, SaveTask, linearly_connect_tasks, MergeFeatureTask
from eolearn.io import SentinelHubDemTask, SentinelHubEvalscriptTask, SentinelHubInputTask
CLIENT_ID = ""
CLIENT_SECRET = ""
config = SHConfig()
if CLIENT_ID and CLIENT_SECRET:
config.sh_client_id = CLIENT_ID
config.sh_client_secret = CLIENT_SECRET
if config.sh_client_id == "" or config.sh_client_secret == "" or config.instance_id == "":
print("Warning! To use Sentinel Hub services, please provide the credentials (client ID and client secret).")
bands_evalscript = """
//VERSION=3
function setup() {
return {
input: [{
bands: [
"ThermalInfrared1", "dataMask"
]
}],
output: {
id: "ms_data",
bands: 2,
sampleType: "FLOAT32"
}
}
}
function evaluatePixel(samples) {
return [samples.ThermalInfrared1, samples.dataMask]
}
"""
eopatch_path = "/home/nedhorning/Abe/TestDataForJohn/EOLearn_Testing/"
epsg = "EPSG:32618"
time_interval = ("2023-05-01", "2023-5-10")
resolution = 30
add_indices = SentinelHubEvalscriptTask(
features=[(FeatureType.DATA, "ms_data")],
evalscript=bands_evalscript,
data_collection=DataCollection.HARMONIZED_LANDSAT_SENTINEL,
resolution=resolution,
aux_request_args={"dataFilter": {"constellation": "LANDSAT"}},
config=config,
max_threads=3
)
roi_bbox = BBox([656839, 4981014, 657406, 4981528], crs=CRS(epsg))
save = SaveTask(eopatch_path + "numpyHLS_Landsat/", overwrite_permission=2, compress_level=0)
workflow_nodes = linearly_connect_tasks(add_indices, save)
workflow = EOWorkflow(workflow_nodes)
result = workflow.execute(
{
workflow_nodes[0]: {"bbox": roi_bbox, "time_interval": time_interval},
workflow_nodes[-1]: {"eopatch_folder": 'test'},
}
)