Hi @rsm278 ,
evalscript.js:46: TypeError: ColorGradientVisualizer.createblueRedTemperature is not a function
This error means that createblueRedTemperature in not a pre-defined function. All pre-defined functions can be found here. If you wish to have your own color scale, the ColorRampVisualizer will do the job. For example, the following script can generate a color ramp from red to blue in a range of 200 to 300.
const ramps = [
[200, 0xff0000],
[300, 0x0000ff ],
];
const visualizer = new ColorRampVisualizer(ramps);
visualizer.process(199); // [ 1, 0, 0 ]
visualizer.process(200); // [ 1, 0, 0 ]
visualizer.process(250); // [ 0.5019607843137255, 0, 0.5019607843137255 ]
visualizer.process(299); // [ 0.011764705882352941, 0, 0.9882352941176471 ]
visualizer.process(300); // [ 0, 1, 0 ]
Regarding the question marks in the commented lines, I believe the one at line 12 is °C as you can see in the script at line 110 the degree Kelvin is converted to the degree Celsius. The other question mark at line 8 is probably an error caused by format or font. I will do a double check.
At line 139, 146, and 148 I get warnings but errors. In general, it does not crash the script but you can do the following to avoid those warnings.
// calc final stdev value
for (var j=0;j<LSTarray.length;j++) {
LSTstd=LSTstd+(Math.pow(LSTarray[j]-LSTavg, 2));
}
LSTstd=(Math.pow(LSTstd/(LSTarray.length-1), 0.5));
// WHICH LST to output, it depends on option variable: 0 for one image analysis (OE Browser); MULTI-TEMPORAL: 0->avg; 1->max; 2->stdev
let outLST= (option==0) ? LSTavg : (option==1) ? LSTmax : LSTstd;
The output is always the same because the default mosaicking is SIMPLE, which flattens the mosaicked image so only a single sample is passed to evaluation (only one value is returned so the average and maximum are the same). To make the options work you have to specify mosaicking: "ORBIT" under the setup() function and remove the square brackets at line 147.
Here’s an example:
//VERSION=3 (auto-converted from 1)
//// STARTING OPTIONS
// choose on basis of which band (B10 or B11) LST mapping will be done
var band="B10";
// for analysis of one image (OE Browser), choose option=0. In case of MULTI-TEMPORAL analyis, option values are following:
// 0 - outputs average LST in selected timeline (% of cloud coverage should be low, e.g. < 10%)
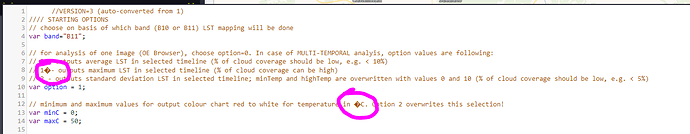
// 1�- outputs maximum LST in selected timeline (% of cloud coverage can be high)
// 2 - outputs standard deviation LST in selected timeline; minTemp and highTemp are overwritten with values 0 and 10 (% of cloud coverage should be low, e.g. < 5%)
var option =2;
// minimum and maximum values for output colour chart red to white for temperature in �C. Option 2 overwrites this selection!
var minC = 0;
var maxC = 50;
////INPUT DATA - FOR BETTER RESULTS, THE DATA SHOULD BE ADJUSTED
// NVDIs for bare soil and NDVIv for full vegetation
// Note: NVDIs for bare soil and NDVIv for full vegetation are needed to be evaluated for every scene. However in the custom script, default values are set regarding:
// http://profhorn.meteor.wisc.edu/wxwise/satmet/lesson3/ndvi.html
// https://www.researchgate.net/post/Can_anyone_help_me_to_define_a_range_of_NDVI_value_to_extract_bare_soil_pixels_for_Landsat_TM
// NVDIs=0.2, NDVIv=0.8
// other source suggests global values: NVDIs=0.2, NDVIv=0.5; https://www.researchgate.net/publication/296414003_Algorithm_for_Automated_Mapping_of_Land_Surface_Temperature_Using_LANDSAT_8_Satellite_Data
var NDVIs=0.2;
var NDVIv=0.8;
// emissivity
var waterE=0.991;
var soilE=0.966;
var vegetationE=0.973;
//var buildingE=0.962;
var C=0.009; //surface roughness, https://www.researchgate.net/publication/331047755_Land_Surface_Temperature_Retrieval_from_LANDSAT-8_Thermal_Infrared_Sensor_Data_and_Validation_with_Infrared_Thermometer_Camera
//central/mean wavelength in meters, B10 or B11
var bCent= (band=="B10") ? 0.000010895 : 0.000012005;
// rho =h*c/sigma=PlanckC*velocityLight/BoltzmannC
var rho=0.01438; // m K
//// visualization
// if result should be std dev (option=2), overwrite minMaxC.
if (option==2) {
minC = 0;
maxC = 25;
}
let viz = ColorGradientVisualizer.createRedTemperature(minC, maxC);
function setup() {
return {
input: [{
bands: [
"B03",
"B04",
"B05",
"B10",
"B11"
]
}],
output: { bands: 3 },
mosaicking: "ORBIT"
}
}
//emissivity calc
//https://www.researchgate.net/publication/296414003_Algorithm_for_Automated_Mapping_of_Land_Surface_Temperature_Using_LANDSAT_8_Satellite_Data
//https://www.academia.edu/27239873/Investigating_Land_Surface_Temperature_Changes_Using_Landsat_Data_in_Konya_Turkey
function LSEcalc(NDVI,Pv) {
var LSE;
if (NDVI < 0) {
//water
LSE=waterE;
} else if (NDVI < NDVIs) {
//soil
LSE=soilE;
} else if (NDVI > NDVIv) {
//vegetation
LSE=vegetationE;
} else {
//mixtures of vegetation and soil
LSE=vegetationE*Pv+soilE*(1-Pv)+C;
}
return LSE;
}
function evaluatePixelOrig(samples) {
// starting values max, avg, stdev, reduce N, N for multi-temporal
var LSTmax=-999;
var LSTavg=0;
var LSTstd=0;
var reduceNavg=0;
var N=samples.length;
//to caputure all values of one pixel for for whole timeline in mosaic order
var LSTarray = [];
// multi-temporal: loop all samples in selected timeline
for (var i=0;i<N;i++) {
//// for LST
// B10 or B11
var Bi= (band=="B10") ? samples[i].B10 : samples[i].B11;
var B03i = samples[i].B03;
var B04i = samples[i].B04;
var B05i = samples[i].B05;
// some images have errors, whole area is either B10<173K or B10>65000K. Also errors, where B04 and B05 =0. Therefore no processing if that happens, in addition for average and stdev calc, N has to be reduced!
if ((Bi>173 && Bi<65000) && (B03i>0 && B04i>0 && B05i>0)) {
// ok image
//1 Kelvin to C
var b10BTi=Bi-273.15;
//2 NDVI - Normalized Difference vegetation Index
var NDVIi=(B05i-B04i)/(B05i+B04i);
//3 PV - proportional vegetation
var PVi=Math.pow(((NDVIi-NDVIs)/(NDVIv-NDVIs)), 2);
//4 LSE land surface emmisivity
var LSEi=LSEcalc(NDVIi,PVi);
//5 LST
var LSTi=(b10BTi/(1+(((bCent*b10BTi)/rho)*Math.log(LSEi))));
////temporary calculation
//avg
LSTavg=LSTavg+LSTi;
//max
if (LSTi>LSTmax) {LSTmax=LSTi;}
//array
LSTarray.push(LSTi);
} else {
// image NOT ok
++reduceNavg;
}
}
// correct N value if some images have errors and are not analysed
N=N-reduceNavg;
// calc final avg value
LSTavg=LSTavg/N;
// calc final stdev value
for (var j=0;j<LSTarray.length;j++) {
LSTstd=LSTstd+(Math.pow(LSTarray[j]-LSTavg, 2));
}
LSTstd=(Math.pow(LSTstd/(LSTarray.length-1), 0.5));
// WHICH LST to output, it depends on option variable: 0 for one image analysis (OE Browser); MULTI-TEMPORAL: 0->avg; 1->max; 2->stdev
let outLST= (option==0)? LSTavg : (option==1) ? LSTmax : LSTstd;
//// output to image
return viz.process(outLST);
}
function evaluatePixel(sample, scene, metadata, customData, outputMetadata) {
return evaluatePixelOrig(sample, scene, metadata, customData, outputMetadata);
}
Note: Legend can only be added with the plotting package, e.g. matplotlib for Python. You may want to remove the visualiser in the script to return actual values, so you can render a color map and have the legend with it.