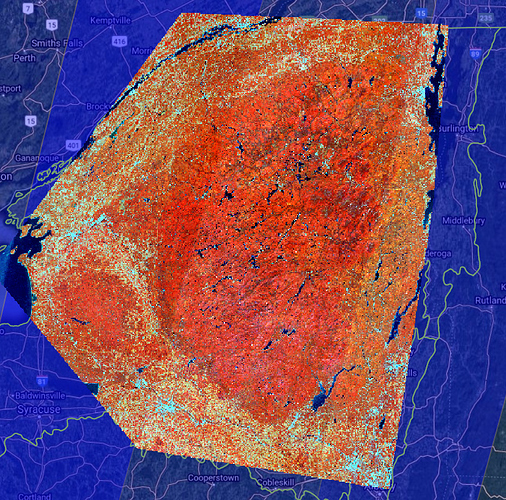
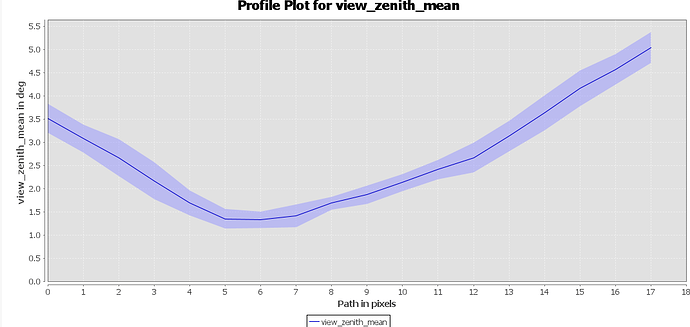
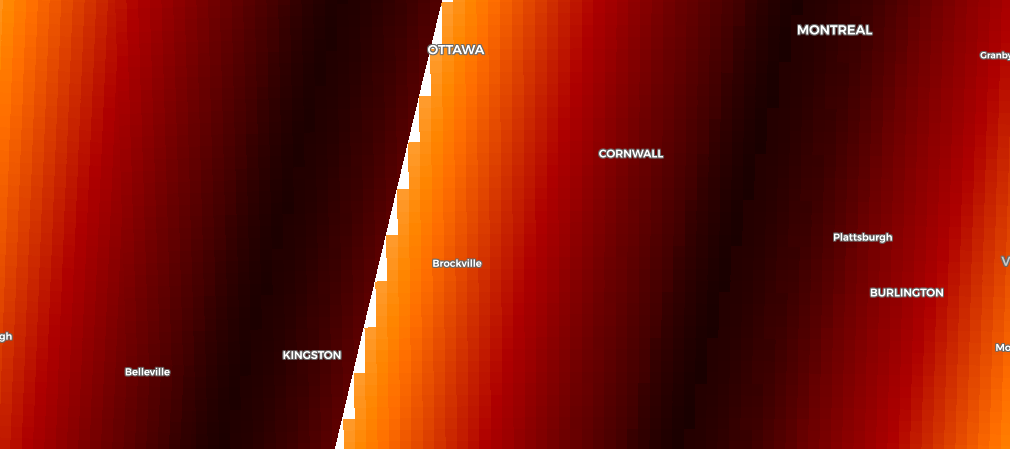
I am attempting to create large mosaics from Sentinel-2 for modeling vegetation structure. I have set up a custom script to create 20 meter resolution 5-band (2, 3, 4, 8, 11) cloudless mosaics filtered down to collections that occur during the summer months (June/July/August) of 2018, 2019, and 2020. This script is performing well in many places but there are cases where the orbit patterns are clearly visible in the output image. Here is a low-resolution preview that shows the problematic pattern:
The nominal sentinel 2 orbit paths are displayed beneath the mosaic image, the darker blue areas are overlap zones between orbits. The stripe on the eastern edge of the mosaic is the most problematic, the source images appear to have very different spectral characteristics (lighter tones on eastern side, darker tones on western side).
Are there tweaks parameters in the Evalscript or in the request (i.e. date ranges, mosaicking settings, etc) that might help reduce the between-orbit stripe artifacts?
Input data request:
"data": [
{
"type": "S2L2A",
"dataFilter": {
"timeRange": {
"from": "2018-06-15T00:00:00Z",
"to": "2020-09-01T23:59:59Z"
},
"maxCloudCoverage": 50,
"mosaickingOrder": "mostRecent"
}
}
]
Evalscript:
//VERSION=3
// copied from here:
// https://github.com/sentinel-hub/custom-scripts/blob/master/sentinel-2/cloudless_mosaic/L2A-first_quartile_4bands.js
function setup() {
return {
input: [{
bands: [
"B04", // red
"B03", // green
"B02", // blue
"B08", // nir
"B11", // swir
"SCL" // pixel classification
],
units: "DN"
}],
output: {
bands: 5,
sampleType: SampleType.UINT16
},
mosaicking: "ORBIT"
};
}
// acceptable images are ones collected in summer months of 2018, 2019, 2020
function filterScenes(availableScenes, inputMetadata) {
return availableScenes.filter(function(scene) {
var m = scene.date.getMonth();
var y = scene.date.getFullYear();
var years = [2018, 2019, 2020];
var months = [5, 6, 7]; // 0 indexed, 5 = June, 6 = July, 7 = August
return months.includes(m) && years.includes(y);
});
}
function getValue(values) {
values.sort(function (a, b) {
return a - b;
});
return getMedian(values);
}
// function for pulling first quartile of values
function getFirstQuartile(sortedValues) {
var index = Math.floor(sortedValues.length / 4);
return sortedValues[index];
}
// function for pulling median (second quartile) of values
function getMedian(sortedValues) {
var index = Math.floor(sortedValues.length / 2);
return sortedValues[index];
}
function validate(samples) {
var scl = samples.SCL;
if (scl === 3) { // SC_CLOUD_SHADOW
return false;
} else if (scl === 9) { // SC_CLOUD_HIGH_PROBA
return false;
} else if (scl === 8) { // SC_CLOUD_MEDIUM_PROBA
return false;
} else if (scl === 7) { // SC_CLOUD_LOW_PROBA
// return false;
} else if (scl === 10) { // SC_THIN_CIRRUS
return false;
} else if (scl === 11) { // SC_SNOW_ICE
return false;
} else if (scl === 1) { // SC_SATURATED_DEFECTIVE
return false;
} else if (scl === 2) { // SC_DARK_FEATURE_SHADOW
// return false;
}
return true;
}
function evaluatePixel(samples, scenes) {
var clo_b02 = [];
var clo_b03 = [];
var clo_b04 = [];
var clo_b08 = [];
var clo_b11 = [];
var clo_b02_invalid = [];
var clo_b03_invalid = [];
var clo_b04_invalid = [];
var clo_b08_invalid = [];
var clo_b11_invalid = [];
var a = 0;
var a_invalid = 0;
for (var i = 0; i < samples.length; i++) {
var sample = samples[i];
if (sample.B02 > 0 && sample.B03 > 0 && sample.B04 > 0 && sample.B08 > 0 && sample.B11 > 0) {
var isValid = validate(sample);
if (isValid) {
clo_b02[a] = sample.B02;
clo_b03[a] = sample.B03;
clo_b04[a] = sample.B04;
clo_b08[a] = sample.B08;
clo_b11[a] = sample.B11;
a = a + 1;
} else {
clo_b02_invalid[a_invalid] = sample.B02;
clo_b03_invalid[a_invalid] = sample.B03;
clo_b04_invalid[a_invalid] = sample.B04;
clo_b08_invalid[a_invalid] = sample.B08;
clo_b11_invalid[a_invalid] = sample.B11;
a_invalid = a_invalid + 1;
}
}
}
var rValue;
var gValue;
var bValue;
var nValue;
var sValue;
if (a > 0) {
rValue = getValue(clo_b04);
gValue = getValue(clo_b03);
bValue = getValue(clo_b02);
nValue = getValue(clo_b08);
sValue = getValue(clo_b11);
} else if (a_invalid > 0) {
rValue = getValue(clo_b04_invalid);
gValue = getValue(clo_b03_invalid);
bValue = getValue(clo_b02_invalid);
nValue = getValue(clo_b08_invalid);
sValue = getValue(clo_b11_invalid);
} else {
rValue = 0;
gValue = 0;
bValue = 0;
nValue = 0;
sValue = 0;
}
return [rValue, gValue, bValue, nValue, sValue];
}