Hello sentinelhub-Team,
I downloaded Sentinel-2 B04 & B08 (NDVI) and Sentinel-1 VH & VV-data.
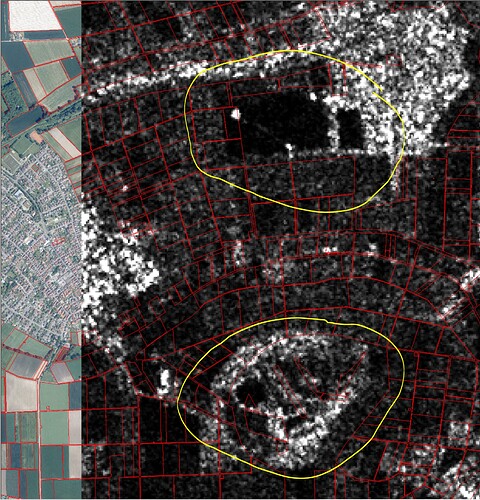
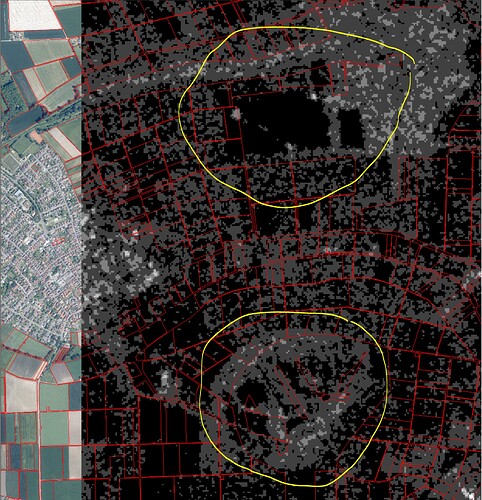
The Sentinel-2 data ist geometrical correct with my ground truth data.
The Sentinel-1-data has geometric error/geometric offset to the ground truth data with depends from EOpatch from 100 up to 400 m.
The scripts are following. How is the geometric offset posible?
Sentinel-1-script:
# Firstly, some necessary imports
# Jupyter notebook related
%reload_ext autoreload
%autoreload 2
%matplotlib inline
import datetime
##import itertools
# Built-in modules
import os
# Basics of Python data handling and visualization
import numpy as np
##from aenum import MultiValueEnum
##np.random.seed(42)
import geopandas as gpd
##import joblib
# Machine learning
##import lightgbm as lgb
import matplotlib.pyplot as plt
##from matplotlib.colors import BoundaryNorm, ListedColormap
from shapely.geometry import Polygon
##from sklearn import metrics, preprocessing
from tqdm.auto import tqdm
from sentinelhub import DataCollection, UtmZoneSplitter
# Imports from eo-learn and sentinelhub-py
from eolearn.core import (
EOExecutor,
EOPatch,
EOTask,
EOWorkflow,
FeatureType,
LoadTask,
MergeFeatureTask,
OverwritePermission,
SaveTask,
linearly_connect_tasks,
)
##from eolearn.features import LinearInterpolationTask, NormalizedDifferenceIndexTask, SimpleFilterTask
##from eolearn.geometry import ErosionTask, VectorToRasterTask
from eolearn.io import ExportToTiffTask, SentinelHubInputTask, VectorImportTask
##from eolearn.ml_tools import FractionSamplingTask
#Get country boundary
# Folder where data for running the notebook is stored
#Daten-Ordner
DATA_FOLDER = os.path.join("/Sen1_Radar_Download/") #/Sen1_Radar_Download/Hessen_diss_utmWgs84_32632.geojson
#EOPatch-Folder
EOPATCH_FOLDER = os.path.join(".", "Sen1_Radar_Download", "EO20230305_VH")
print ("EOPATCH_FOLDER: ", EOPATCH_FOLDER)
# Locations for collected data and intermediate results
#Test-Ordner
EOPATCH_SAMPLES_FOLDER = os.path.join(".", "eopatches_sampled")
#Ergebnis
RESULTS_FOLDER = DATA_FOLDER
for folder in (EOPATCH_FOLDER, EOPATCH_SAMPLES_FOLDER, RESULTS_FOLDER):
os.makedirs(folder, exist_ok=True)
# Load geojson file
#country = gpd.read_file(os.path.join(DATA_FOLDER, "svn_border.geojson"))
#country = gpd.read_file(os.path.join(DATA_FOLDER, "Hessen_diss_utmWgs84_32632.geojson"))
country = gpd.read_file(os.path.join(DATA_FOLDER, "NEWGEB_Wolle_Wgs84_32632.geojson"))
print ("Country: ", country )
# Add 500m buffer to secure sufficient data near border
country = country.buffer(500)
# Get the country's shape in polygon format
country_shape = country.geometry.values[0]
# Plot country
country.plot()
plt.axis("off")
# Print size
country_width = country_shape.bounds[2] - country_shape.bounds[0]
country_height = country_shape.bounds[3] - country_shape.bounds[1]
print(f"Dimension of the area is {country_width:.0f} x {country_height:.0f} m2")
#Split to smaller tiles and choose a 5x5 area
# Create a splitter to obtain a list of bboxes with 5km sides
bbox_splitter = UtmZoneSplitter([country_shape], country.crs, 5000)
bbox_list = np.array(bbox_splitter.get_bbox_list())
info_list = np.array(bbox_splitter.get_info_list())
# Prepare info of selected EOPatches
geometry = [Polygon(bbox.get_polygon()) for bbox in bbox_list]
idxs = [info["index"] for info in info_list]
idxs_x = [info["index_x"] for info in info_list]
idxs_y = [info["index_y"] for info in info_list]
print ("idxs: ", idxs, "idxs_x: ", idxs_x, "idxs_y: ", idxs_y )
bbox_gdf = gpd.GeoDataFrame({"index": idxs, "index_x": idxs_x, "index_y": idxs_y}, crs=country.crs, geometry=geometry)
# select a 5x5 area (id of center patch)
ID = 27#00
# Obtain surrounding 5x5 patches
patchIDs = []
for idx, (bbox, info) in enumerate(zip(bbox_list, info_list)):
if abs(info["index_x"] - info_list[ID]["index_x"]) <= 2 and abs(info["index_y"] - info_list[ID]["index_y"]) <= 2:
patchIDs.append(idx)
# Check if final size is 5x5
if len(patchIDs) != 5 * 5:
print("Warning! Use a different central patch ID, this one is on the border.")
# Change the order of the patches (useful for plotting)
patchIDs = np.transpose(np.fliplr(np.array(patchIDs).reshape(5, 5))).ravel()
# Save to shapefile
shapefile_name = "grid_NewGebWolle_500x500.gpkg"
bbox_gdf.to_file(os.path.join(RESULTS_FOLDER, shapefile_name), driver="GPKG")
#Visualize the selection
# Display bboxes over country
fig, ax = plt.subplots(figsize=(30, 30))
ax.set_title("Selected 5x5 tiles from Wolle NO-Rheinobergraben", fontsize=25)
country.plot(ax=ax, facecolor="w", edgecolor="b", alpha=0.5)
bbox_gdf.plot(ax=ax, facecolor="w", edgecolor="r", alpha=0.5)
for bbox, info in zip(bbox_list, info_list):
geo = bbox.geometry
ax.text(geo.centroid.x, geo.centroid.y, info["index"], ha="center", va="center")
# Mark bboxes of selected area
bbox_gdf[bbox_gdf.index.isin(patchIDs)].plot(ax=ax, facecolor="g", edgecolor="r", alpha=0.5)
plt.axis("off");
#Inputtask
# BAND DATA
# Add a request for S2 bands.
# Here we also do a simple filter of cloudy scenes (on tile level).
# The s2cloudless masks and probabilities are requested via additional data.
band_names = ["Radar"]
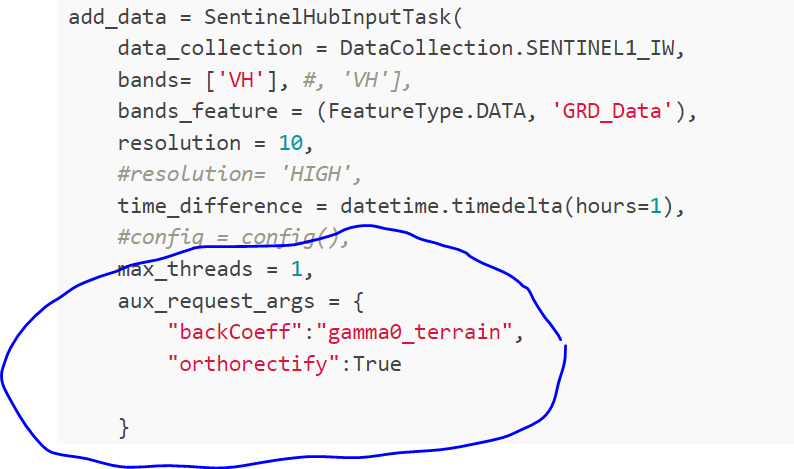
add_data = SentinelHubInputTask(
data_collection = DataCollection.SENTINEL1_IW,
bands= ['VH'], #, 'VH'],
bands_feature = (FeatureType.DATA, 'GRD_Data'),
resolution = 10,
#resolution= 'HIGH',
time_difference = datetime.timedelta(hours=1),
#config = config(),
max_threads = 1,
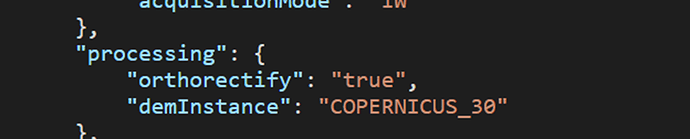
aux_request_args = {
"backCoeff":"gamma0_terrain",
"orthorectify":True
}
)
# VALIDITY MASK
# Validate pixels using SentinelHub's cloud detection mask and region of acquisition
add_sh_validmask = SentinelHubValidDataTask((FeatureType.MASK, "IS_VALID"))
# COUNTING VALID PIXELS
# Count the number of valid observations per pixel using valid data mask
add_valid_count = AddValidCountTask("IS_VALID", "VALID_COUNT")
# SAVING TO OUTPUT (if needed)
save = SaveTask(EOPATCH_FOLDER, overwrite_permission=OverwritePermission.OVERWRITE_PATCH)
# Define the workflow
workflow_nodes = linearly_connect_tasks(
add_data, save
)
workflow = EOWorkflow(workflow_nodes)
# Let's visualize it
workflow.dependency_graph()
print ("patchIDs", patchIDs)
for idx, bbox in enumerate(bbox_list[patchIDs]):
print ("idx: ", idx, "bbox: ", bbox,"patchIDs", patchIDs[idx])
%%time
# Time interval for the SH request
time_interval = ["2018-04-01", "2018-10-31"]
# Define additional parameters of the workflow
input_node = workflow_nodes[0]
save_node = workflow_nodes[-1]
execution_args = []
for idx, bbox in enumerate(bbox_list[patchIDs]):
print ("patchIDs[idx]", patchIDs[idx])
execution_args.append(
{
input_node: {"bbox": bbox, "time_interval": time_interval},
save_node: {"eopatch_folder": f"eopatch_{patchIDs[idx]}"},
}
)
# Execute the workflow
executor = EOExecutor(workflow, execution_args, save_logs=True)
executor.run(workers=4)
executor.make_report()
failed_ids = executor.get_failed_executions()
if failed_ids:
raise RuntimeError(
f"Execution failed EOPatches with IDs:\n{failed_ids}\n"
f"For more info check report at {executor.get_report_path()}"
)
Download Sentinel-2 B04 B08 → NDVI
# Firstly, some necessary imports
# Jupyter notebook related
%reload_ext autoreload
%autoreload 2
%matplotlib inline
import datetime
#import itertools
# Built-in modules
import os
# Basics of Python data handling and visualization
import numpy as np
#from aenum import MultiValueEnum
np.random.seed(42)
import geopandas as gpd
#import joblib
# Machine learning
#import lightgbm as lgb
import matplotlib.pyplot as plt
#from matplotlib.colors import BoundaryNorm, ListedColormap
from shapely.geometry import Polygon
#from sklearn import metrics, preprocessing
from tqdm.auto import tqdm
from sentinelhub import DataCollection, UtmZoneSplitter
# Imports from eo-learn and sentinelhub-py
from eolearn.core import (
EOExecutor,
EOPatch,
EOTask,
EOWorkflow,
FeatureType,
LoadTask,
MergeFeatureTask,
OverwritePermission,
SaveTask,
linearly_connect_tasks,
)
#from eolearn.features import LinearInterpolationTask, NormalizedDifferenceIndexTask, SimpleFilterTask
#from eolearn.geometry import ErosionTask, VectorToRasterTask
from eolearn.io import ExportToTiffTask, SentinelHubInputTask, VectorImportTask
#from eolearn.ml_tools import FractionSamplingTask
# Folder where data for running the notebook is stored
DATA_FOLDER = os.path.join("/Sen1_Radar_Download/") #/Sen1_Radar_Download/Hessen_diss_utmWgs84_32632.geojson
EOPATCH_FOLDER = os.path.join(".", "Sen1_Radar_Download", "EO20230305_NDVI_CC0_2")
print ("EOPATCH_FOLDER: ", EOPATCH_FOLDER)
# Locations for collected data and intermediate results
#EOPATCH_FOLDER = os.path.join(".", "eopatches")
EOPATCH_SAMPLES_FOLDER = os.path.join(".", "eopatches_sampled")
#RESULTS_FOLDER = os.path.join(".", "results")
RESULTS_FOLDER = DATA_FOLDER
for folder in (EOPATCH_FOLDER, EOPATCH_SAMPLES_FOLDER, RESULTS_FOLDER):
os.makedirs(folder, exist_ok=True)
# Load geojson file
country = gpd.read_file(os.path.join(DATA_FOLDER, "NEWGEB_Wolle_Wgs84_32632.geojson"))
print ("Country: ", country )
# Add 500m buffer to secure sufficient data near border
country = country.buffer(500)
# Get the country's shape in polygon format
country_shape = country.geometry.values[0]
# Plot country
country.plot()
plt.axis("off")
# Print size
country_width = country_shape.bounds[2] - country_shape.bounds[0]
country_height = country_shape.bounds[3] - country_shape.bounds[1]
print(f"Dimension of the area is {country_width:.0f} x {country_height:.0f} m2")
# Create a splitter to obtain a list of bboxes with 5km sides
bbox_splitter = UtmZoneSplitter([country_shape], country.crs, 5000)
bbox_list = np.array(bbox_splitter.get_bbox_list())
info_list = np.array(bbox_splitter.get_info_list())
# Prepare info of selected EOPatches
geometry = [Polygon(bbox.get_polygon()) for bbox in bbox_list]
idxs = [info["index"] for info in info_list]
idxs_x = [info["index_x"] for info in info_list]
idxs_y = [info["index_y"] for info in info_list]
print ("idxs: ", idxs, "idxs_x: ", idxs_x, "idxs_y: ", idxs_y )
bbox_gdf = gpd.GeoDataFrame({"index": idxs, "index_x": idxs_x, "index_y": idxs_y}, crs=country.crs, geometry=geometry)
# select a 5x5 area (id of center patch)
ID = 27#00
# Obtain surrounding 5x5 patches
patchIDs = []
for idx, (bbox, info) in enumerate(zip(bbox_list, info_list)):
if abs(info["index_x"] - info_list[ID]["index_x"]) <= 2 and abs(info["index_y"] - info_list[ID]["index_y"]) <= 2:
patchIDs.append(idx)
# Check if final size is 5x5
if len(patchIDs) != 5 * 5:
print("Warning! Use a different central patch ID, this one is on the border.")
# Change the order of the patches (useful for plotting)
patchIDs = np.transpose(np.fliplr(np.array(patchIDs).reshape(5, 5))).ravel()
# Save to shapefile
shapefile_name = "grid_NewGebWolle_500x500.gpkg"
bbox_gdf.to_file(os.path.join(RESULTS_FOLDER, shapefile_name), driver="GPKG")
# Display bboxes over country
fig, ax = plt.subplots(figsize=(30, 30))
ax.set_title("Selected 5x5 tiles from Wolle NO-Rheinobergraben", fontsize=25)
country.plot(ax=ax, facecolor="w", edgecolor="b", alpha=0.5)
bbox_gdf.plot(ax=ax, facecolor="w", edgecolor="r", alpha=0.5)
for bbox, info in zip(bbox_list, info_list):
geo = bbox.geometry
ax.text(geo.centroid.x, geo.centroid.y, info["index"], ha="center", va="center")
# Mark bboxes of selected area
bbox_gdf[bbox_gdf.index.isin(patchIDs)].plot(ax=ax, facecolor="g", edgecolor="r", alpha=0.5)
plt.axis("off");
class SentinelHubValidDataTask(EOTask):
"""
Combine Sen2Cor's classification map with `IS_DATA` to define a `VALID_DATA_SH` mask
The SentinelHub's cloud mask is asumed to be found in eopatch.mask['CLM']
"""
def __init__(self, output_feature):
self.output_feature = output_feature
def execute(self, eopatch):
eopatch[self.output_feature] = eopatch.mask["IS_DATA"].astype(bool) & (~eopatch.mask["CLM"].astype(bool))
return eopatch
class AddValidCountTask(EOTask):
"""
The task counts number of valid observations in time-series and stores the results in the timeless mask.
"""
def __init__(self, count_what, feature_name):
self.what = count_what
self.name = feature_name
def execute(self, eopatch):
eopatch[FeatureType.MASK_TIMELESS, self.name] = np.count_nonzero(eopatch.mask[self.what], axis=0)
return eopatch
# BAND DATA
# Add a request for S2 bands.
# Here we also do a simple filter of cloudy scenes (on tile level).
# The s2cloudless masks and probabilities are requested via additional data.
#band_names = ["Radar"]
band_names = ['B04', 'B08']
#band_names = ['PSI', 'NDVI2']
add_data = SentinelHubInputTask(
bands_feature=(FeatureType.DATA, 'BANDS'),
bands = band_names,
resolution=10,
maxcc=0.2,
time_difference=datetime.timedelta(minutes=120),
data_collection =DataCollection.SENTINEL2_L1C,
additional_data=[(FeatureType.MASK, 'dataMask', 'IS_DATA'),
(FeatureType.MASK, 'CLM'),
(FeatureType.DATA, 'CLP')],
max_threads=5
)
ndvi = NormalizedDifferenceIndexTask((FeatureType.DATA, 'BANDS'), (FeatureType.DATA, 'NDVI'),
[band_names.index('B08'), band_names.index('B04')])
# VALIDITY MASK
# Validate pixels using SentinelHub's cloud detection mask and region of acquisition
add_sh_validmask = SentinelHubValidDataTask((FeatureType.MASK, "IS_VALID"))
# COUNTING VALID PIXELS
# Count the number of valid observations per pixel using valid data mask
add_valid_count = AddValidCountTask("IS_VALID", "VALID_COUNT")
# SAVING TO OUTPUT (if needed)
save = SaveTask(EOPATCH_FOLDER, overwrite_permission=OverwritePermission.OVERWRITE_PATCH)
# Define the workflow
workflow_nodes = linearly_connect_tasks(
add_data,
ndvi,
add_sh_validmask,
add_valid_count,
save
)
workflow = EOWorkflow(workflow_nodes)
# Let's visualize it
workflow.dependency_graph()
%%time
# Time interval for the SH request
time_interval = ["2018-04-01", "2018-10-31"]
# Define additional parameters of the workflow
input_node = workflow_nodes[0]
save_node = workflow_nodes[-1]
execution_args = []
for idx, bbox in enumerate(bbox_list[patchIDs]):
print ("patchIDs[idx]", patchIDs[idx])
execution_args.append(
{
input_node: {"bbox": bbox, "time_interval": time_interval},
save_node: {"eopatch_folder": f"eopatch_{patchIDs[idx]}"},
}
)
# Execute the workflow
executor = EOExecutor(workflow, execution_args, save_logs=True)
executor.run(workers=4)
executor.make_report()
failed_ids = executor.get_failed_executions()
if failed_ids:
raise RuntimeError(
f"Execution failed EOPatches with IDs:\n{failed_ids}\n"
f"For more info check report at {executor.get_report_path()}"
)