Hi @s_practice ,
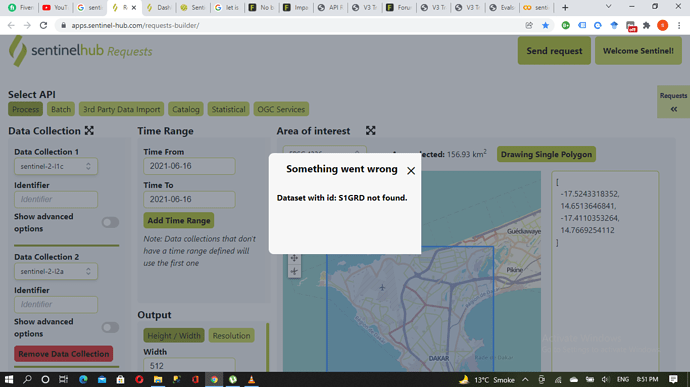
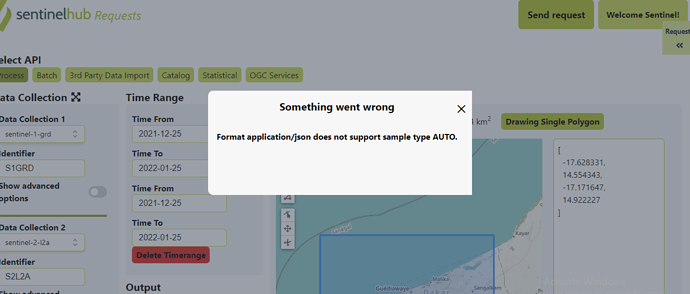
The Sand-Oriented Land Cover Classification script uses Sentinel Hub Data Fusion capabilities. Since we are requesting data from two different data sources (Sentinel-1 and Sentinel-2) we need to add separable identifiers to the API call in the Evalscript and the request payload.
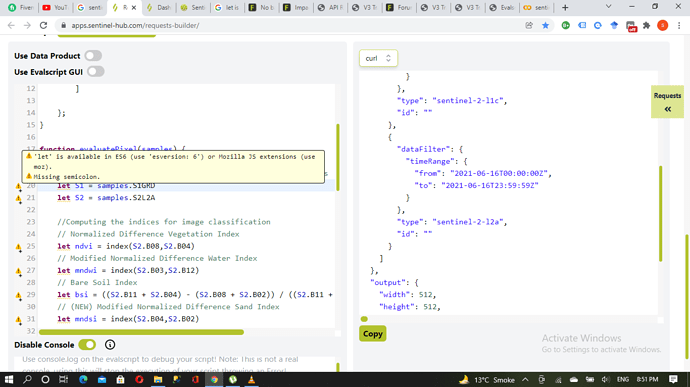
In the Evalscript, you can see that the respective Sentinel-1 and Sentinel-2 bands are assigned to the identifiers S1GRD and S2L2A.
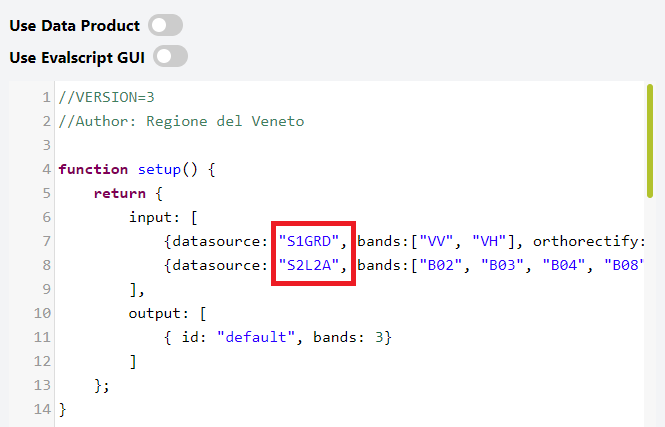
Note that it’s up to you which strings you want to use as identifiers here, as long as they correspond to the identifiers you specify in the request payload!
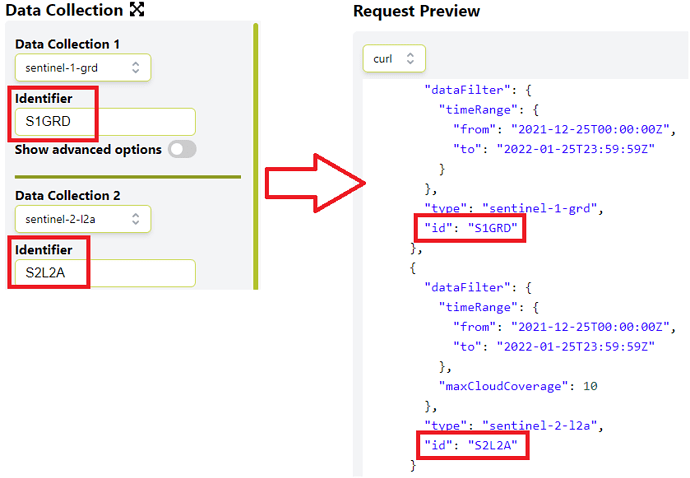
So if you add S1GRD and S2L2A to the respective identifier field in the Data Collection section, the Requests Builder will automatically add the two identifiers to the request payload as can be seen in the next screenshot, and your request should run without a problem.
If you plan to use the functionality more regularly, you should find everything you need in our dedicated documentation section.
Here is a working request example you can copy and paste into the Request preview window:
curl -X POST https://services.sentinel-hub.com/api/v1/process
-H 'Content-Type: application/json'
-H 'Authorization: Bearer <your access token>'
-d '{
"input": {
"bounds": {
"bbox": [
-17.628331,
14.554343,
-17.171647,
14.922227
]
},
"data": [
{
"dataFilter": {
"timeRange": {
"from": "2021-12-25T00:00:00Z",
"to": "2022-01-25T23:59:59Z"
}
},
"type": "sentinel-1-grd",
"id": "S1GRD"
},
{
"dataFilter": {
"timeRange": {
"from": "2021-12-25T00:00:00Z",
"to": "2022-01-25T23:59:59Z"
},
"maxCloudCoverage": 10
},
"type": "sentinel-2-l2a",
"id": "S2L2A"
}
]
},
"output": {
"width": 512,
"height": 426.118,
"responses": [
{
"identifier": "default",
"format": {
"type": "image/jpeg"
}
}
]
},
"evalscript": "//VERSION=3\n//Author: Regione del Veneto\n\nfunction setup() {\n return {\n input: [\n {datasource: \"S1GRD\", bands:[\"VV\", \"VH\"], orthorectify:\"TRUE\"},\n {datasource: \"S2L2A\", bands:[\"B02\", \"B03\", \"B04\", \"B08\",\"B11\",\"B12\"]}\n ],\n output: [\n { id: \"default\", bands: 3}\n ]\n };\n}\n\nfunction evaluatePixel(samples) {\n\n // Setting the samples for Sentinel-1 and Sentinel-2 products\n\n let S1 = samples.S1GRD[0]\n let S2 = samples.S2L2A[0]\n \n //Computing the indices for image classification\n\n // Normalized Difference Vegetation Index\n\n let ndvi = index(S2.B08,S2.B04)\n // Modified Normalized Difference Water Index\n\n let mndwi = index(S2.B03,S2.B12)\n // Bare Soil Index\n\n let bsi = ((S2.B11 + S2.B04) - (S2.B08 + S2.B02)) / ((S2.B11 + S2.B04) + (S2.B08 + S2.B02))\n // (NEW) Modified Normalized Difference Sand Index\n\n let mndsi = index(S2.B04,S2.B02)\n\n //Setting threshold values for image classification\n\n // Extracts built-up areas in white\n\n if (S1.VH > 0.2 || S1.VV > 0.2) {\n return {\n default: [1,1,1]\n }}\n // Extracts shrub and grassland in ~yellow\n\n if (ndvi > 0.2 && ndvi < 0.4) {\n return {\n default: [0.9,0.9,0]\n }}\n // Extracts flourishing vegetation in ~green\n\n if (ndvi >= 0.4) {\n return {\n default: [0.2,0.75,0]\n }}\n // Extracts water bodies in ~blue\n\n if (mndwi > 0 && S1.VH < 0.015) {\n return {\n default: [0,0,0.75]\n }}\n // Extracts bare soils in ~red\n\n if (bsi > 0.2) {\n return {\n default: [0.75,0,0]\n }}\n // Extracts sand surface in ~orange\n\n if (mndsi > 0.1) {\n return {\n default: [1,0.65,0]\n }}\n // Leave in black unclassified pixels\n\n else{\n return {\n default: [0,0,0]\n }}\n}"
}'
If you then click the Parse button underneath, the Requests Builder mask should be updated with the parameter settings from the posted request.