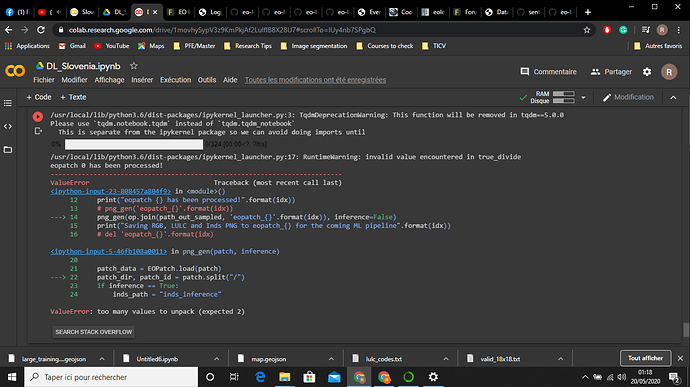
I am trying to follow this tutorial: land cover fastai
And while executing the workflow, I got this error:
This is the code where the error occurs:
def color_scale(arr):
"""correct the RGB bands to be a composed bands of value between 0 255 for visualization purpos
Args:
arr: RGB bands in numpy array
Return:
str_arr: numpy array that values range from 0 to 255 for visualization
"""
str_arr = (arr + 1) * 127.5
return str_arr
def png_gen(patch, inference=False):
"""Save RGB, spectral info and labeled images for the coming deep learning
Args:
patch: Saved eopatches for deep learning LULC training and prediction
Return:
None: images in the PNG.
"""
patch_data = EOPatch.load(patch)
patch_dir, patch_id = patch.split("/")
if inference == True:
inds_path = "inds_inference"
if not op.isdir(inds_path):
makedirs(inds_path)
for i in range(len(bands)):
#get NDVI, NDWI and NDBI from the bands
inds = color_scale(bands[i][..., [-3, -2, -1]]).astype("uint8")
Image.fromarray(inds).save(op.join(inds_path, patch+'_'+str(i)+'.png'))
else:
lulc = patch_data.mask_timeless['LULC']
lulc_path = "lulc_all"
rgb_path = "rgb_all"
inds_path = "inds_all"
if not op.isdir(lulc_path):
makedirs(lulc_path)
if not op.isdir(rgb_path):
makedirs(rgb_path)
if not op.isdir(inds_path):
makedirs(inds_path)
for i in range(len(bands)):
# Switch R, G, B band index from bands
rgb = color_scale(bands[i][..., [2, 1, 0]]).astype("uint8")
inds = color_scale(bands[i][..., [-3, -2, -1]]).astype("uint8")
Image.fromarray(rgb).save(op.join(rgb_path, "{}_{}.png".format(patch_id, str(i))))
Image.fromarray(inds).save(op.join(inds_path, "{}_{}.png".format(patch_id, str(i))))
Image.fromarray(lulc).save(op.join(lulc_path, "{}_{}.png".format(patch_id, str(i))))
And this is the execution code:
#download eopatch for the desired AOI and covert the numpy array into RGB, spectral info (NDVI, NDWI, NDBI)
# and training lulc label into PNG and saved them under "rgb_all", "inds_all" and "lulc_all"
pbar = tqdm(total=len(patchIDs))
for idx, bbox in enumerate(tile_list[patchIDs]):
# define additional parameters of the workflow
extra_param = {
add_data: {'bbox': bbox, 'time_interval': time_interval},
save_s2: {'eopatch_folder': 'eopatch_{}'.format(idx)}
}
workflow.execute(extra_param)
print("eopatch {} has been processed!".format(idx))
# png_gen('eopatch_{}'.format(idx))
png_gen(op.join(path_out_sampled, 'eopatch_{}'.format(idx)), inference=False)
print("Saving RGB, LULC and Inds PNG to eopatch_{} for the coming ML pipeline".format(idx))
# del 'eopatch_{}'.format(idx)
shutil.rmtree(op.join(path_out_sampled, 'eopatch_{}'.format(idx)), ignore_errors=True)
print("eopatch_{} deleted!".format(idx))
pbar.update(1)
Instead of ‘/’ I tried ‘_’ in this line patch_dir, patch_id = patch.split("/") because the downloaded patches have this format “eopatch_idx”, but I got the same error.